Algorithms and Computational Biology Lab
Department of Computer Science and Engineering
Engineering Building Unit II, Room 362
University of California
Riverside CA 92521
I will be working as a Principal Scientist II at Roche Sequencing Solutions, Pleasanton, CA starting December, 2019.
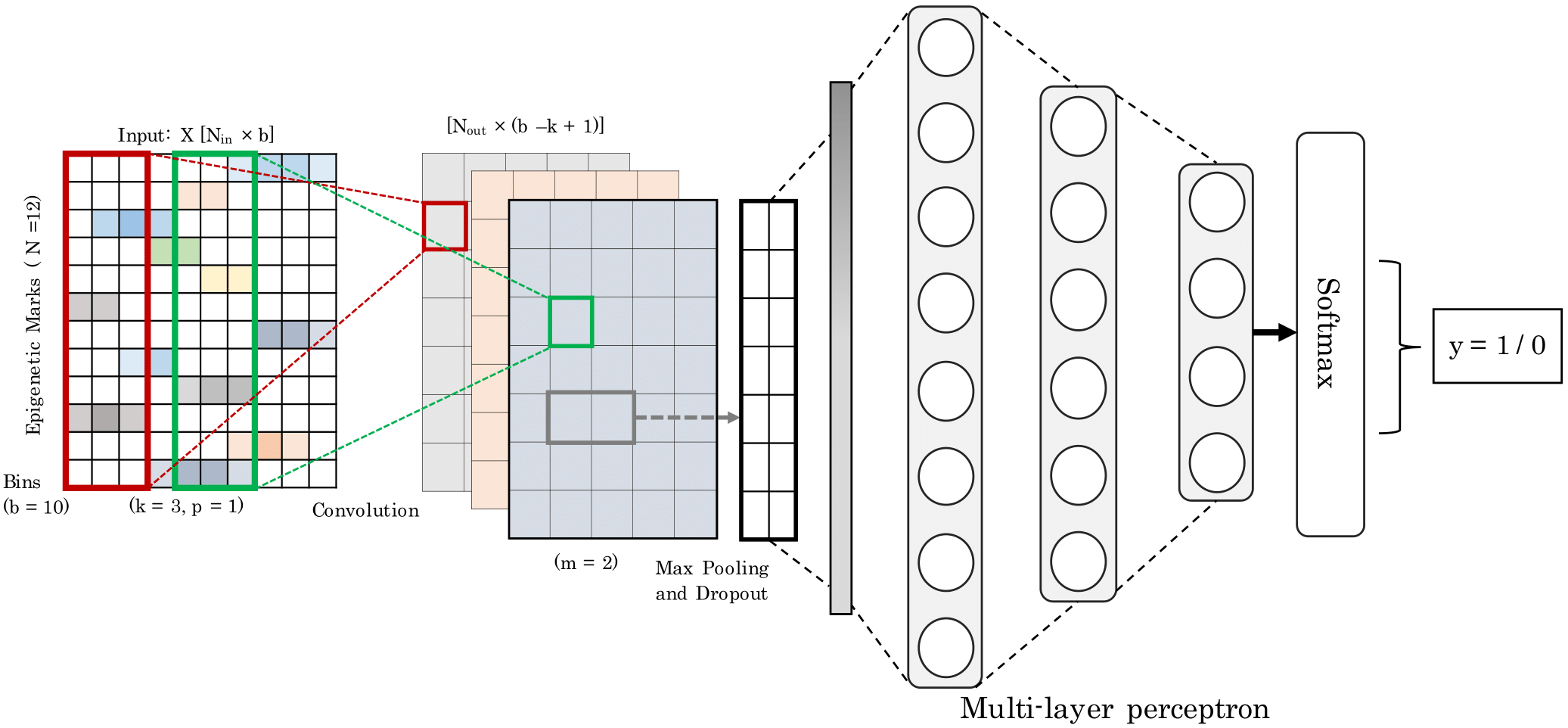
Enhancers are distal cis-regulatory elements that play a critical role in gene expression control. Their remote location relative to the target gene(s), their complex functional properties, and their lack of discriminating motifs and evolutionary-conserved sequence makes it challenging to detect them. The availability of genome-wide epigenetics data for a variety of cell lines has opened the possibility for a machine learning approach for the identification of enhancer regions. We propose a convolutional neural network called Epi2En for the prediction of enhancers across multiple human cell-lines from twelve epigenetic features.
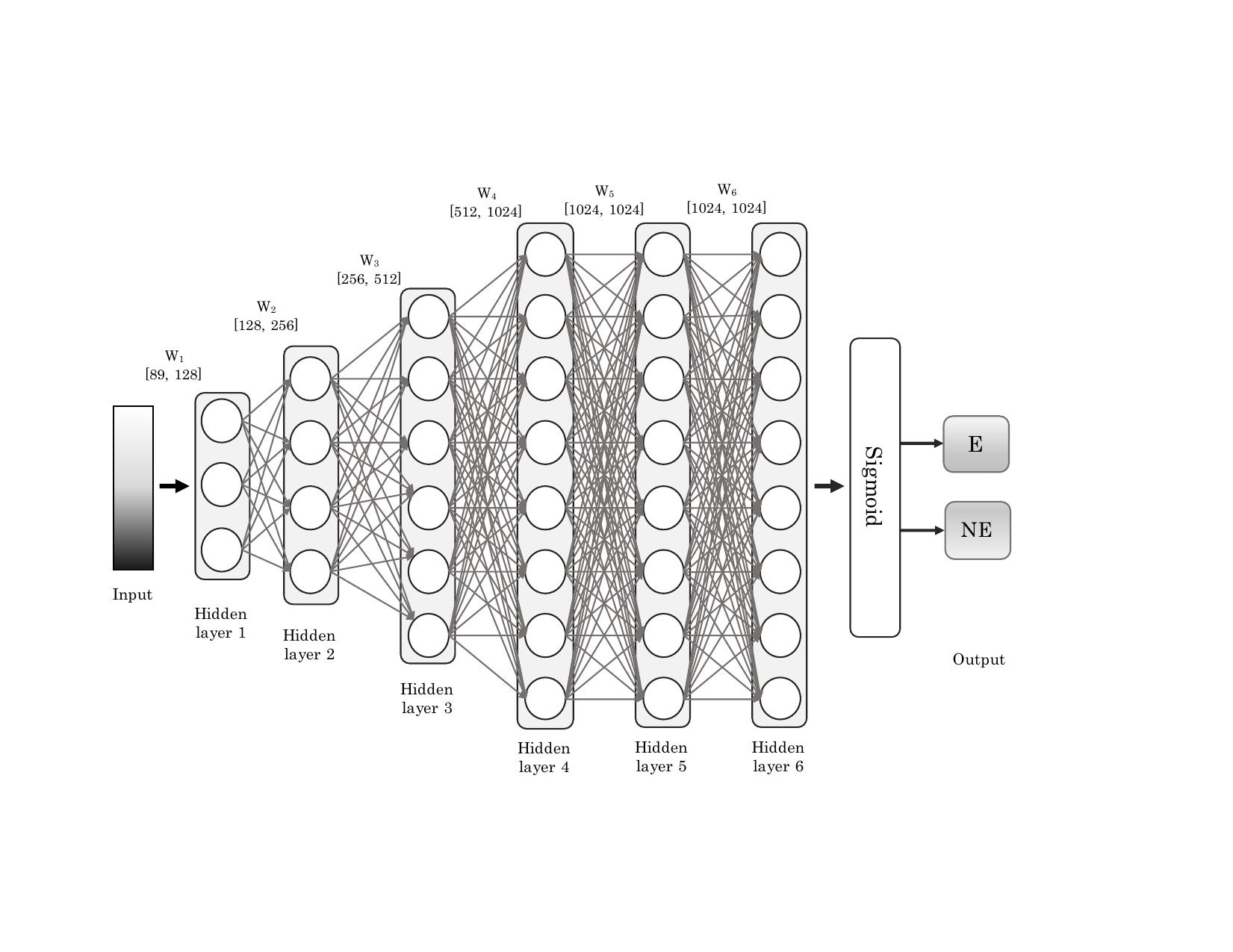
Essential genes are critical for the survival of an organism. The prediction of essential genes in bacteria can provide targets for the design of novel antibiotic compounds or antimicrobial strategies. We have built a deep neural network classifier (DeeplyEssential) to classify essential and non-essential genes in these bacteria species. Our model only uses sequence-based features to predict the gene essentiality. Moreover we also expose and study a hidden performance bias that affected the previous classifier.
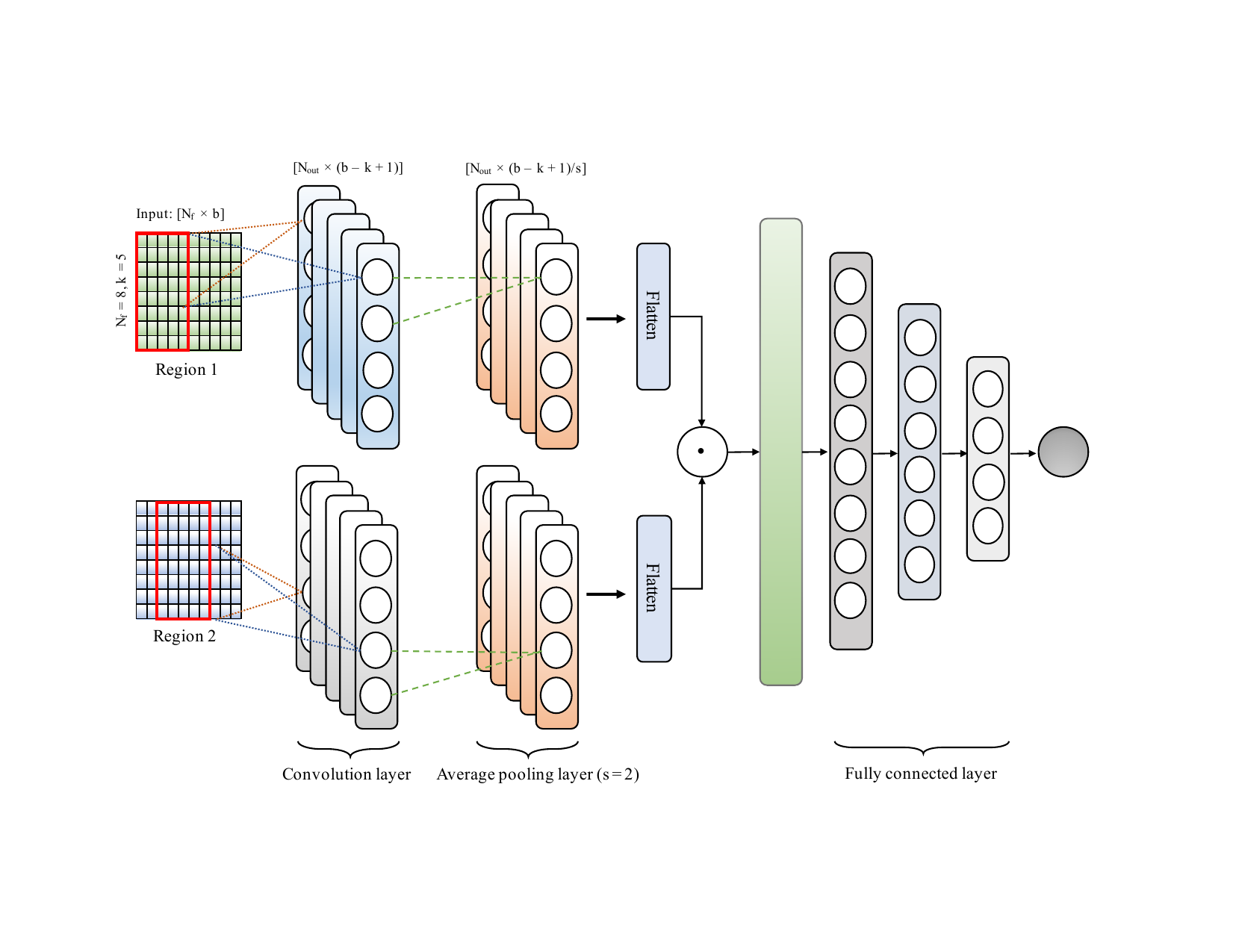
On this ongoing project, we are trying to predict HiC interaction between two regions using epigenetic signals. HiC interaction can have major roles in tissue-specific expression and how regulatory sequence variants impact complex phenotype including diseases such as cancer diabetes and obesity. There are very few HiC datasets available at high resolution since the experiment is very expensive. As a result, the computational approach for predicting long-range interaction.
In this work, we are building feature matrices from the epigenetic signals which then processes by a convolutional neural network for predicting the interaction between two regions. In this work, we will be investigating combinatorial relationships among features, and the ability of the predicted interactions to identify, topologically associated domains (TADs), loops, contact counts in different cell lines etc.
September 2014 - November 2019
Supervisor: Prof. Stefano Lonardi
December 2010 - December 2012
Supervisor: Prof. M. A. Mottalib
January 2006 - November 2010