DAME :
Disk-Aware Motif
Enumeration
This is a supporting page to our paper -
A Disk-Aware Algorithm for Time Series Motif Discovery, by Abdullah Mueen, Eamonn Keogh and Nima Bigdely-Shamlo
The paper is here.
This is a supporting page to our paper -
A Disk-Aware Algorithm for Time Series Motif Discovery, by Abdullah Mueen, Eamonn Keogh and Nima Bigdely-Shamlo
The paper is here.
Gunnar Waterstraat from CharitÚ - Universitńtsmedizin Berlin has made a linux version of the code which is available here. Thanks to Gunnar Waterstraat.
We
have compiled results of all the experiments in an spreadsheet.
In the subsequent sections, we will refer to sheets from this document.
Execution times may not be exactly the same as in the spreadsheet
because of the random referencing, but they should be very close and
representative for the claim of our paper. It is worth
mentioning some notes about the document.
- All the real datasets are single time series and processed by the damesub.cpp code.
- Locations of motifs are all indexed from 0 as in standard C programming language.
- The directory named "temp" holds the sorted database.
- All the times are in seconds unless specified otherwise.
- All the distances are in z-normalized space.
- All the Brute Force algorithms use "Early Abandonning".
randn('state',[362436069;521288629]);
rootDir = 'u';
for i=1:80
c = random_walk(1024,50000);
save(sprintf('%s\\u%d.txt',rootDir,i),'c','-ascii');
end
rootDir = 'u';
for i=1:80
c = random_walk(1024,50000);
save(sprintf('%s\\u%d.txt',rootDir,i),'c','-ascii');
end
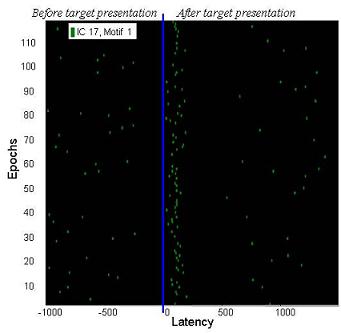
The subsequences that are within the hyperspehere of radius 2 from the Motif 1 have a densed distribution around 100ms after target presentation (shown in the next plot).
In this experiment we use the
database of "80 million
tiny images" from
80 million tiny
images: a large dataset for non-parametric object and scene recognition
by A. Torralba, R. Fergus and W. T. Freeman, IEEE Transactions on Pattern Analysis and Machine Intelligence, vol.30(11), pp. 1958-1970, 2008. For the database please contact with the authors of the above paper.
DAME finds 3,836,902 images which have at least one duplicate in the first 40 million images of the above database. DAME also finds 542,603 images that have at leat one near duplicate in the database with distance less than 0.1. We use tiny2ts.m and im2ts.m to convert the image database into databases of time series of length 768 points. Some representitive duplicates and motifs are presented here. For complete list of duplicates and near-duplicates please email us.
by A. Torralba, R. Fergus and W. T. Freeman, IEEE Transactions on Pattern Analysis and Machine Intelligence, vol.30(11), pp. 1958-1970, 2008. For the database please contact with the authors of the above paper.
DAME finds 3,836,902 images which have at least one duplicate in the first 40 million images of the above database. DAME also finds 542,603 images that have at leat one near duplicate in the database with distance less than 0.1. We use tiny2ts.m and im2ts.m to convert the image database into databases of time series of length 768 points. Some representitive duplicates and motifs are presented here. For complete list of duplicates and near-duplicates please email us.

The trace is overly sampled with 250Hz sample rate and over 8 million data points. We have downdsampled the trace to about 1 million points by a 8:1 sampling ratio. We have found the following motif using DAME which comes from times when the patient went back and forth between arousal and sleep stage1.
The following figure also shows the motif in the original space. This demonstrates that z-normalization removed the offset errors before computing the Euclidean distance, and therefore, motif pair could be so different in their offset from 0.
We have run DAME on this log and found the motif pair shown on the right column. Here the motif length is more than 24 hours (1500 minutes). The days that poped up as motif are the last two days of the first round of the world cup when 8 teams, which had some chance to move to the next round, played their last match in the first round.
Abdullah Mueen
Department of Computer Science and Engineering,
University of California - Riverside.