
Welcome to the MK Motif Discovery Page
This page is build in support of our SDM 2009 paper, A. Mueen, E. Keogh, Q. Zhu, S. Cash & B. Westover (2009). Exact Discovery of Time Series Motifs [pdf]. You can download all the datasets we used in the paper from here. If you want the code, read on.
Time series motifs are pairs of individual time series, or subsequences of a longer time series, which are very similar to each other. Since the formalism of time series motifs in 2002, dozens of researchers have used them in domains as diverse as medicine, entertainment, biology, telemedicine, telepresence and severe weather prediction. Below we show a concrete example.
Consider the time series of insect telemetry below, do you see any reoccurring patterns?

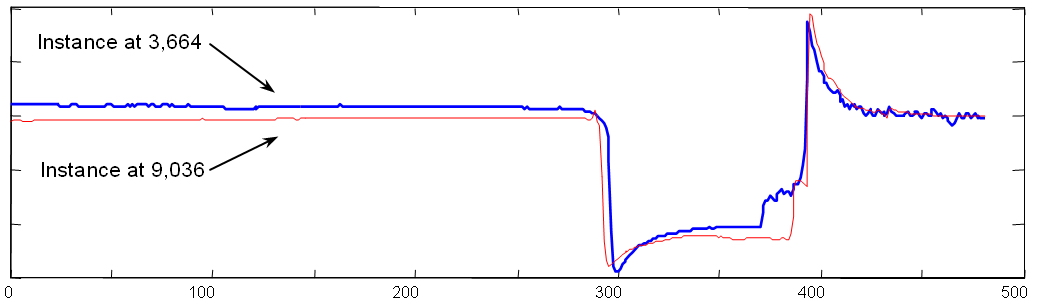
| As it happens, there is a very
interesting repeated pattern as shown to the right (zoomed-in). By
referencing the accompanying video at the relevant locations we are able
to determine that this repeated pattern is not a coincidence, it
corresponds to a behavior that occurs immediately after phloem (plant
sap) ingestion has taken place. This example gives an intuition as to what a time series motif is (see paper for formal details). See this file for more information on the insect problem. |
 |
Naively, using a brute force method, it would take 544,500,000 Euclidean distance calculations to find this motif. Using the MK algorithm, we can reduce this number by several orders of magnitude.
Time series motifs where introduced in our ICDM 2002 paper, and since then there have been dozens of follow up papers. However, MK algorithm is the first non-trivial algorithm to discover exact motifs in large datasets.
Do you want the user-friendly code to find motifs? Download and read this users manual first (powerpoint/pdf), and then download the code.
Acknowledgements: